Investigating patterns with localise
One a bedMethyl table has been created, modkit localise will use the pileup and calculate per-base modification aggregate information around genomic features of interest.
For example, we can investigate base modification patterns around CTCF binding sites.
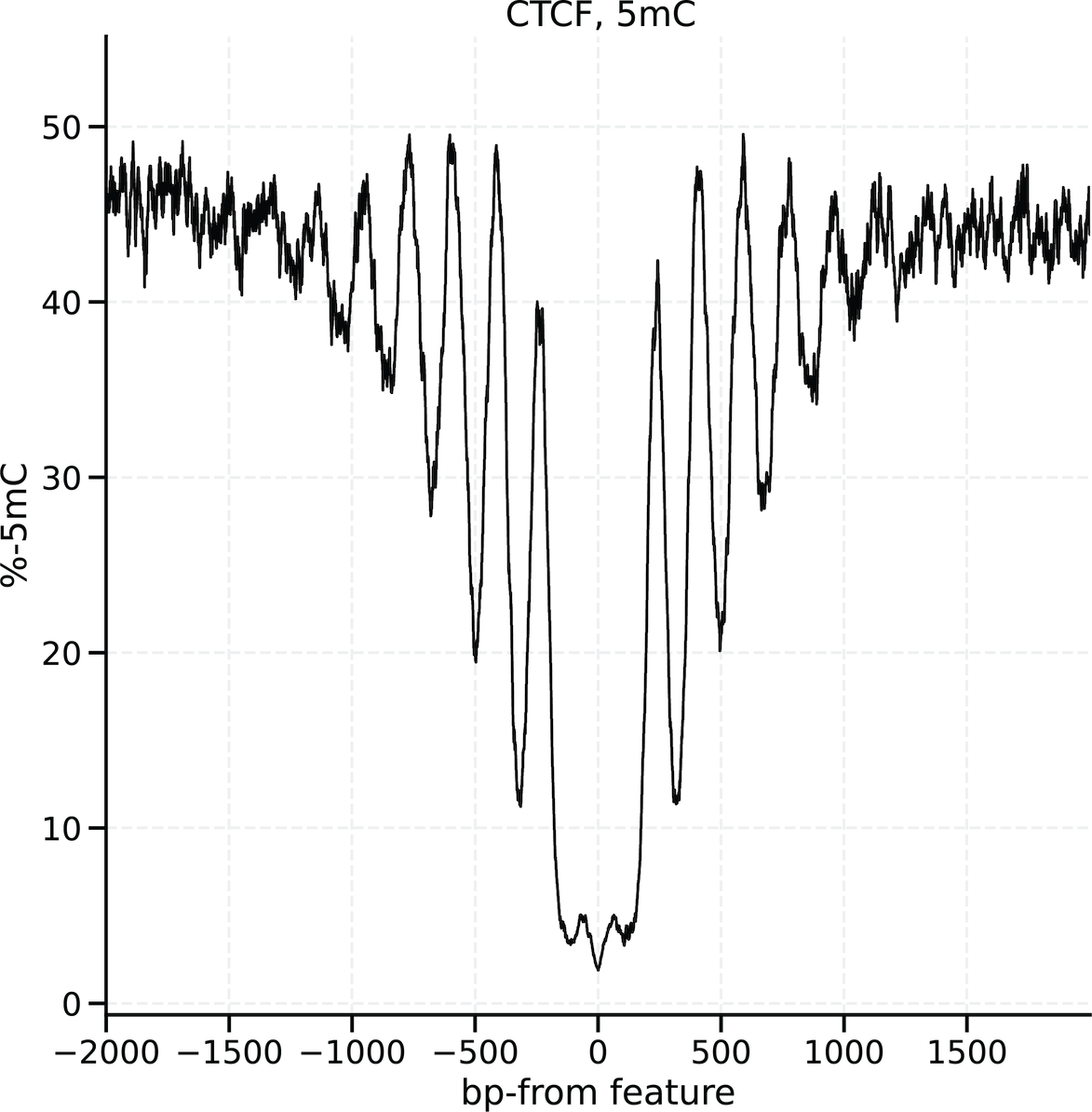
The input requirements to modkit localise are simple:
- BedMethyl table that has been bgzf-compressed and tabix-indexed
- Regions file in BED format (plaintext).
- Genome sizes tab-separated file:
<chrom>\t<size_in_bp>
an example command:
modkit localise ${bedmethyl} --regions ${ctcf} --genome-sizes ${sizes}
The output table has the following schema:
| column | Name | Description | type |
|---|---|---|---|
| 1 | mod code | modification code as present in the bedmethyl | str |
| 2 | offset | distance in base pairs from the center of the genome features, negative values reflect towards the 5’ of the genome | int |
| 3 | n_valid | number of valid calls at this offset for this modification code | int |
| 4 | n_mod | number of calls for this modification code at this offset | int |
| 5 | percent_modified | n_mod / n_valid * 100 | float |
Optionally the --chart argument can be used to create HTML charts of the modification patterns.